miRDeep*
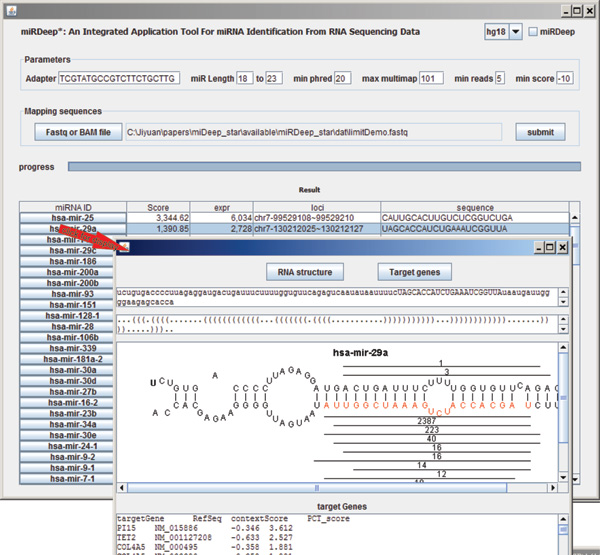
MiRDeep and its varieties are widely used to quantify known and novel miRNA from small RNA sequencing. This paper describes miRDeep*, our integrated miRNA identification tool, which is modeled off miRDeep, but the precision of detecting novel miRNAs is improved by introducing new strategies to identify precursor miRNAs. MiRDeep* has a user-friendly graphic interface and accepts raw data in FastQ and SAM/BAM format. Known and novel miRNA expression levels, as measured by the number of reads, are displayed in an interface which shows each RNAseq read relative to the pre-miRNA hairpin. The secondary pre-miRNA structure and read locations for each predicted miRNA are shown and kept in a separate figure file. Moreover, the target genes of identified known and novel miRNAs are predicted and the targets are ranked according to the confidence score. miRDeep* is an integrated standalone application where sequence alignment, pre-miRNA secondary structure calculation, and graphical display are purely java-coded. This application tool can be executed using a normal personal computer with only 1.5Gb of memory. Using small RNAseq data that was generated in prostate cancer (LNCaP) cells, the precision of miRNA prediction was 83.17%, 81.01% for miRDeep*, compared with 59.14%, 69.23% for miRDeep2, 80.79%, 76.6% for miRDeep, and 19.76%, 20.23% for miRanalyzer. miRDeep* also detected 35-45 novel miRNA compared with 64, 91 for miRDeep2, 39-55 for miRDeep and 848-1060 for miRanalyzer. Our benchmark of miRDeep* against miRDeep2 using a dataset where Dicer was knocked out, demonstrated that miRDeep* is less likely to result in false positive predictions of mature miRNA. Four of the novel miRNA predicted by miRDeep* were validated using stem-loop TaqMan PCR, as well as qualitative RT-PCR targeting the pre-miRNA.
Downloads
new version in sourceforge http://sourceforge.net/projects/mirdeepstar
License
Permission to use this work is granted under the GNU Lesser General Public License (LGPL).
Enquiries
For enquiries about the software please contact Jiyuan An (j.an@qut.edu.au) or Colleen Nelson (colleen.nelson@qut.edu.au)
Tel: +61 7 3443 7280
Related tools
miRDeep, miRDeep2, miRTRAP, MIReNA, miRDeep-P, miRanalyzer. miRNAkey, miRTools, SeqBuster, CPSS, miRGator, targetScan, picard, samtools, miRspring
Citing miRDeep*
An, J., Lai, J., Lehman, M.L. and Nelson, C.C. (2013) miRDeep*: an integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res, 41, 727-737.